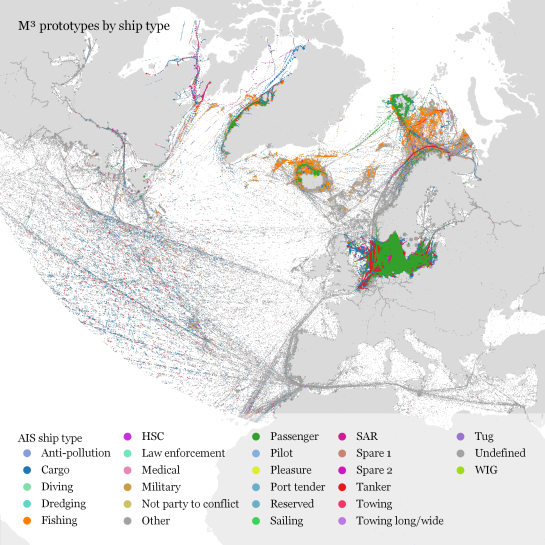
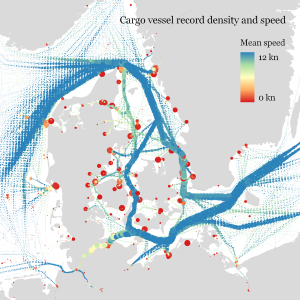
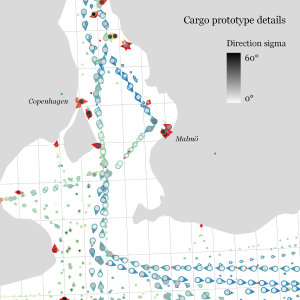
Urban Mobility Insights with MovingPandas & CARTO in Snowflake
Today, I want to point out a blog post over at
https://carto.com/blog/urban-mobility-insights-with-movingpandas-carto-in-snowflake
written together with my fellow co-authors and EMERALDS project team member Argyrios Kyrgiazos.

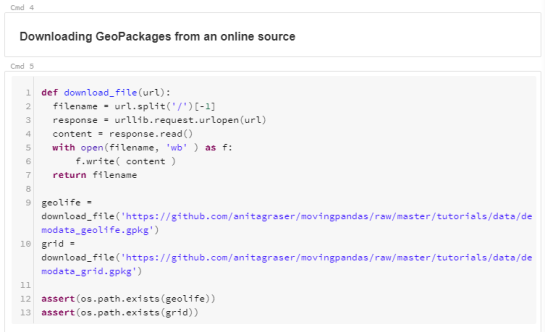
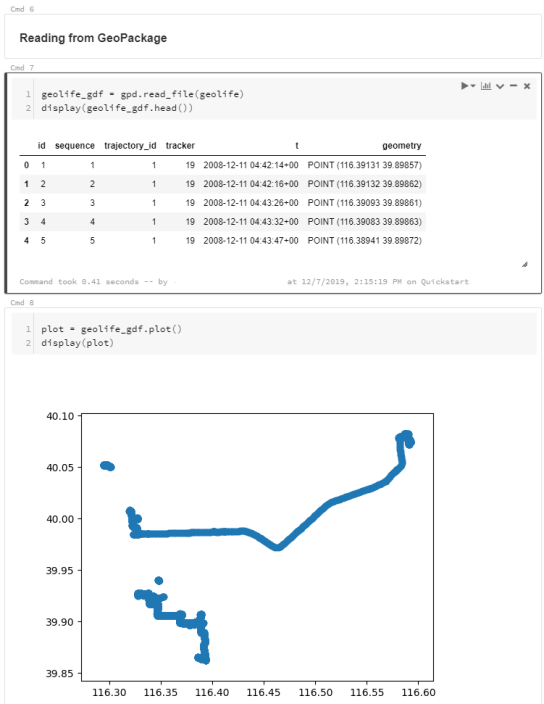
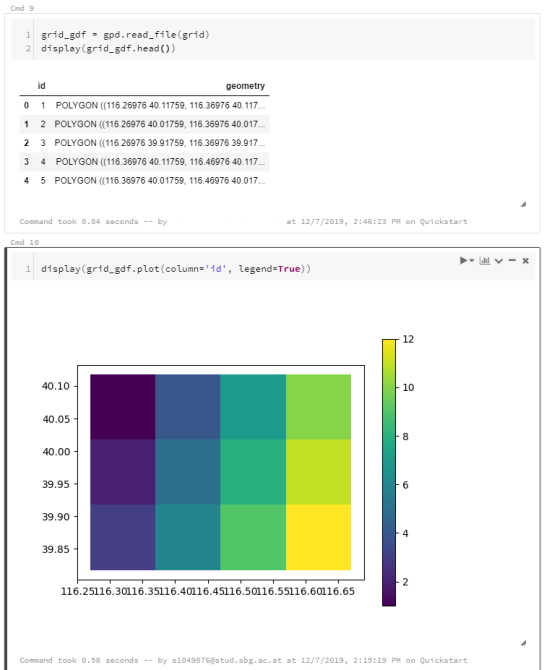
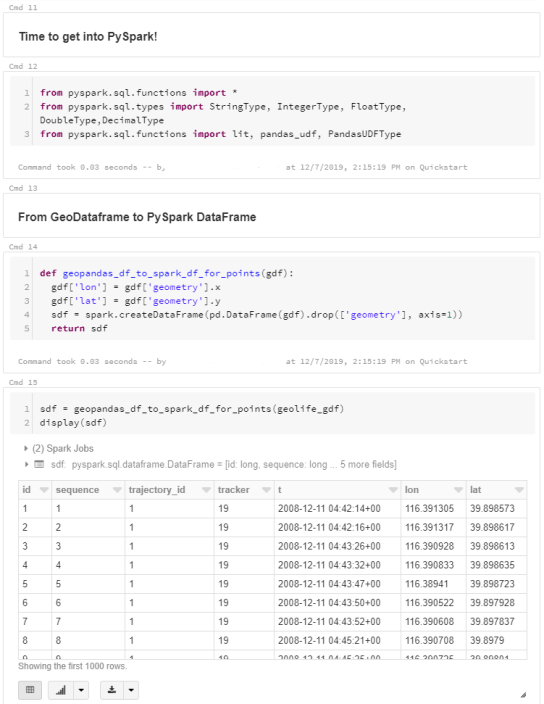
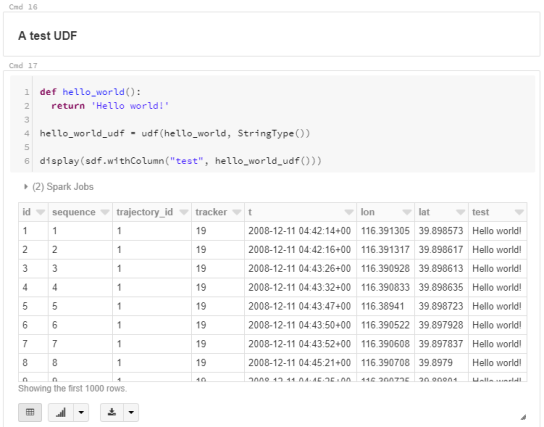
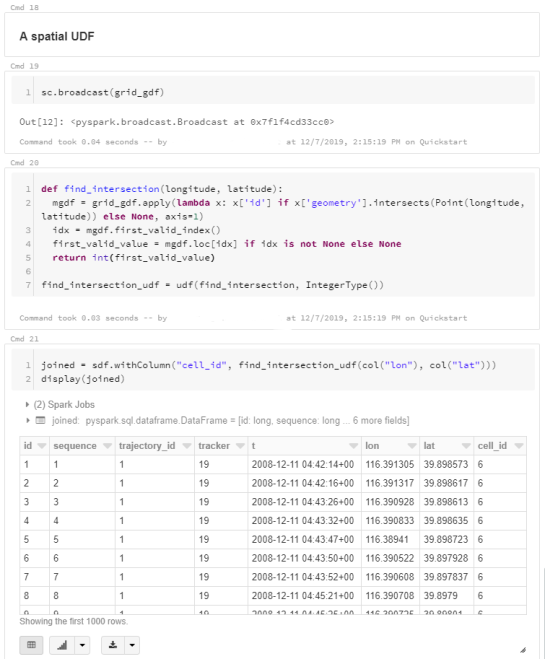
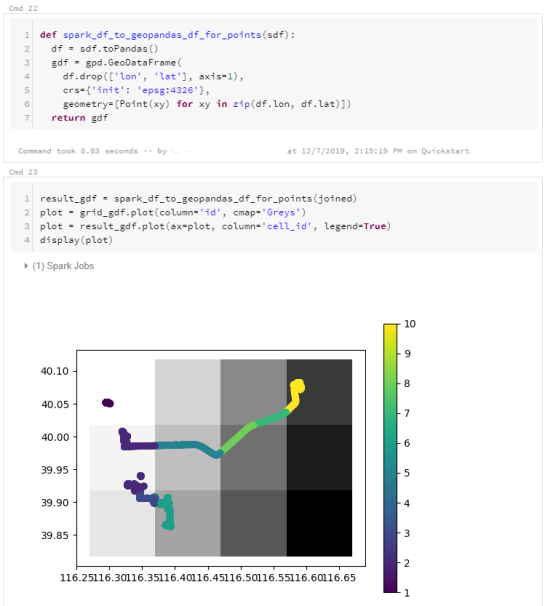
For the technically inclined, the highlight are the presented UDFs in Snowflake to process and transform the trajectory data. For example, here’s a TemporalSplitter UDF:
CREATE OR REPLACE FUNCTION CARTO_DATABASE.CARTO.TemporalSplitter(geom ARRAY, t ARRAY, mode STRING)
RETURNS ARRAY
LANGUAGE PYTHON
RUNTIME_VERSION = 3.11
PACKAGES = ('numpy','pandas', 'geopandas','movingpandas', 'shapely')
HANDLER = 'udf'
AS $$
import numpy as np
import pandas as pd
import geopandas as gpd
import movingpandas as mpd
import shapely
from shapely.geometry import shape, mapping, Point, Polygon
from shapely.validation import make_valid
from datetime import datetime, timedelta
def udf(geom, t, mode):
valid_df = pd.DataFrame(geom, columns=['geometry'])
valid_df['t'] = pd.to_datetime(t)
valid_df['geometry'] = valid_df['geometry'].apply(lambda x:shapely.wkt.loads(x))
gdf = gpd.GeoDataFrame(valid_df, geometry='geometry', crs='epsg:4326')
gdf = gdf.set_index('t')
traj = mpd.Trajectory(gdf, 1)
traj_sm = mpd.TemporalSplitter(traj).split(mode=mode)
if len(traj_sm.trajectories)>0:
res = traj_sm.to_point_gdf()
res['geometry'] = res['geometry'].apply(lambda x: shapely.wkt.dumps(x))
return res.reset_index().values
else:
return []
$$;
You can find the full code here: https://github.com/anitagraser/carto-research-public/tree/master/movingpandas_carto_in_snowflake













 Dash HoloViews was just released as part of
Dash HoloViews was just released as part of  Automatically link selections across multiple plots (crossfiltering)
Automatically link selections across multiple plots (crossfiltering)